Overview
Test blots, as their name implies, are very simple Western blots that are created for the express purpose of optimizing or troubleshooting experimental conditions. They are usually produced by running multiple lanes of the same lysate or purified protein solution on a gel, and after transfer cutting the blot into strips to be tested individually.
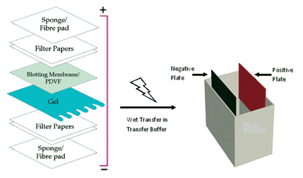
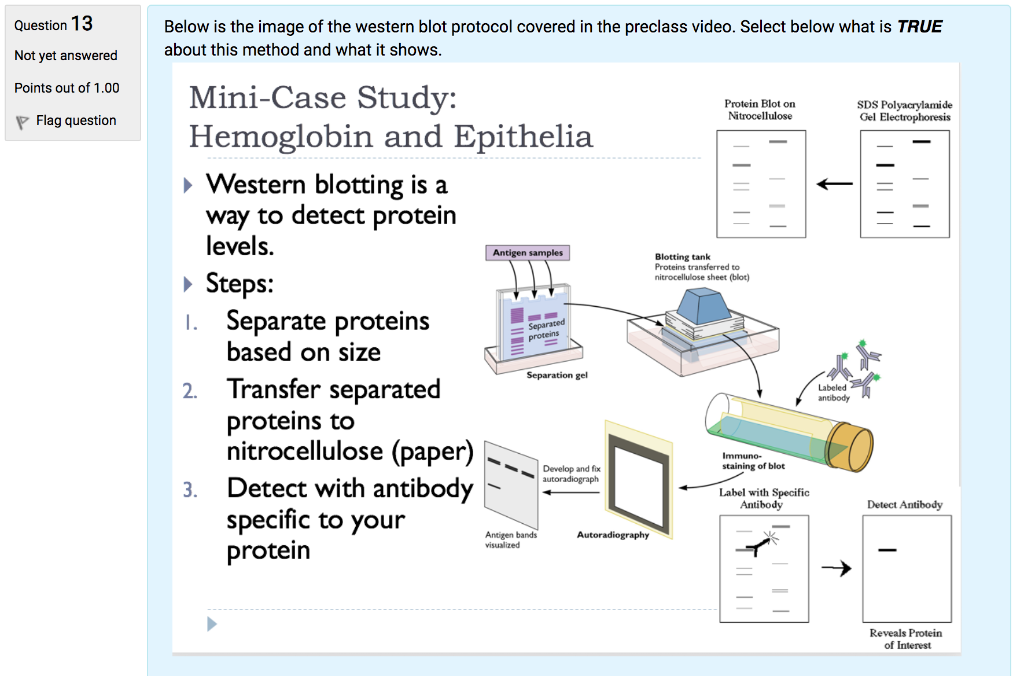
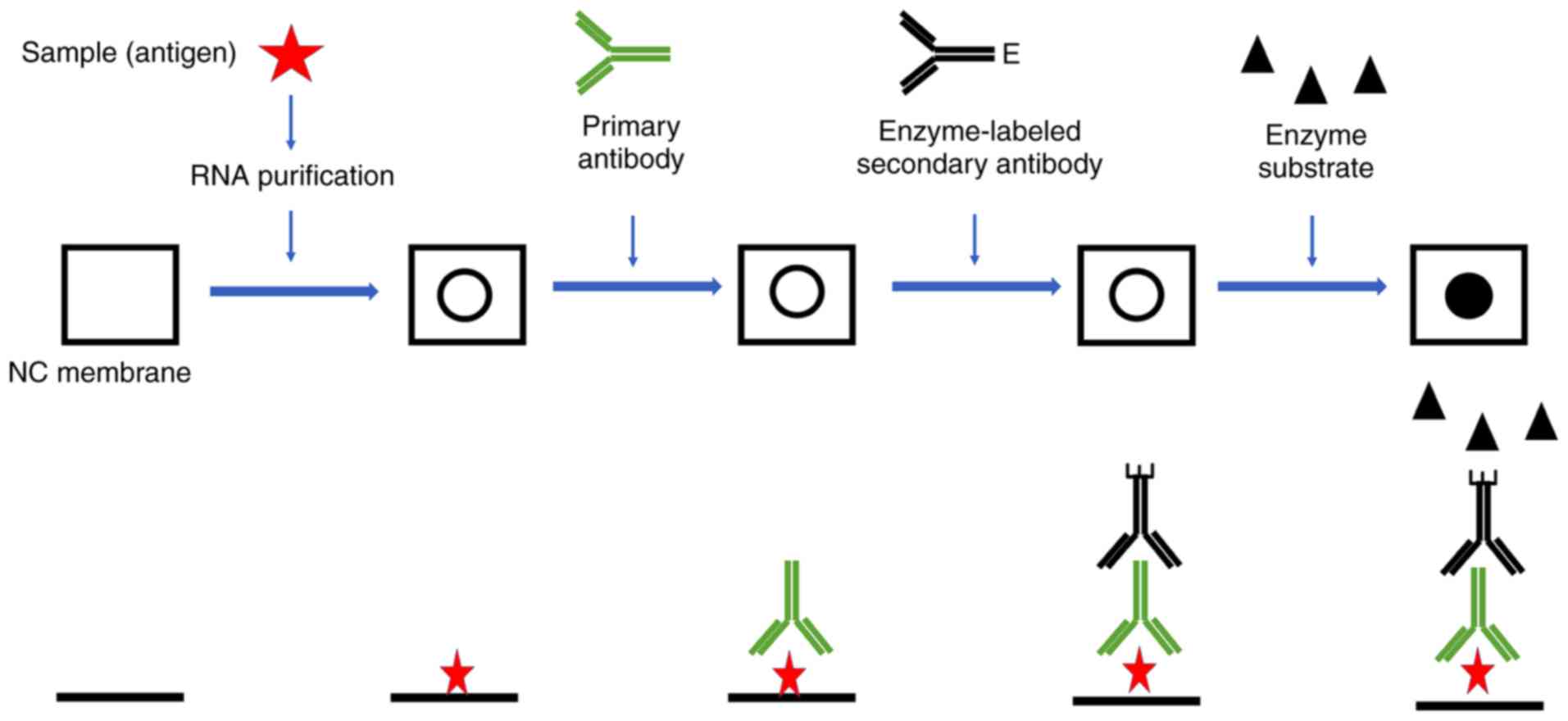
Western blotting of proteins was introduced by Towbin et al. In 1979 and is now a routine and fundamental technique for protein analysis. Western blotting, also called protein blotting or immunoblotting, uses antibodies to identify specific protein targets bound to a membrane; the specificity of the antibody-antigen interaction enables a target protein to be identified in the midst of a. Coats proteins and disrupts secondary and tertiary protein structures by breaking hydrogen bonds and unfolding or denaturing protein. ‘Masks’charge on protein by imparting a large negative charge so that all proteins act the same in regards to charge. Prevents protein aggregation. Prevents protein shape from influencing gel run. Koma protocol If purified, suspend sample in PBS or TBS, or other appropriate buffer. Cell lysates or extracts also may be applied to the membrane or filter for detection of cellular proteins.
They provide a quick and efficient means of examining a range of antibody dilutions or detection substrates
Dot blots and slot blots are also a very useful variation on the typical Western blot. They do not require gel electrophoresis, so there is no separation of proteins by size.
Instead, the target protein or cell lysate mixture is added directly onto the surface of the nitrocellulose or PVDF membrane. Protein solutions can be applied directly in a small volume, or with a vacuum manifold to produce an orderly grid of samples similar to that seen in Figure 14. Each dot or slot blot would contain known amounts of target protein or cell lysate.
Once dry, dot blots and slot blots are subjected to the same immunodetection steps used for Western blotting, i.e. blocking, antibody incubation, and target detection with substrate. Grey and black spots on the figure below indicate which samples are positive for the target protein and correspond roughly to the bands produced on a Western blot.
Figure 14: Dot Blots and Slot Blots. The dot blots at the left represent a dilution series of a sample, with smaller lighter dots corresponding to lower concentrations of target protein. The slot blots represent a group of random samples, the intensity of the signal corresponds to the concentration of the target protein in that sample.
By making a number of identical dots or slots of a single known protein sample, or a range of sample dilutions, one can quickly test several combinations and concentrations of primary and secondary antibodies. Dot blots and slot blots are also beneficial when screening a large number of samples, or if a simple answer will suffice.
Protein Slot Blot Protocol Definition
The main downside to slot blots/dot blots is that they provide no information about molecular weight. Thus, it is harder to detect false positive signals, or to tell whether modified forms of a protein are present.
| Detection with Substrate | Chapter 4: Data Analysis |
This site will not be updated.
homepeopleresearchpublicationsnewsprotocols
Example protocols:
If purified, suspend sample in PBS or TBS, or other appropriate buffer. Cell lysates or extracts also may be applied to the membrane or filter for detection of cellular proteins.If using a filtration manifold, dilute sample to 300-500 µl for a concentration of 1-10 µg/spot, then carry out serial dilutions of the sample to determine the optimal concentrations for subsequent binding assays. If not using a manifold, volumes of 2-5 µl should be used.
If using a filtration manifold to apply samples, float membrane in deionized water until completely wet, then soak in PBS or TBS until use. If spotting without a manifold, membrane should be left dry.
Apply sample to membrane directly with a micropipette, or use a filtration manifold. For purified samples, apply 1-10 µg/spot or well. Sample should be diluted in PBS, TBS or other buffer.
Protein Slot Blot Protocol Igg
Multi-well filtration manifold
Place 2 sheets filter paper, prewet in transfer buffer, on the filter support plate of the Manifold device. Place membrane on top of the filter paper and clamp the sample well plate into place. Apply low vacuum (enough to pull buffer through at approximately 1 mL/minute) and wash each well with 500 µL of sample buffer. While the vacuum is applied, add sample to the well and filter through. After sample has been applied to the membrane, wash with 500 µL of sample buffer. For best results, suspend sample in 300-500 µL of buffer. Unclamp and remove the sample well plate, taking care to leave the membrane on the filter paper. Remove the membrane from the Manifold device.
Direct sample application
If a Manifold device is not used, apply sample to the membrane placed on top of 2 sheets of dry filter paper in 2-5 µL aliquots. If larger sample volumes are used, allow sample spot to dry prior to subsequent 2-5 µL aliquot addition.Note: If using a filtration manifold, low molecular weight samples should be filtered through slowly to prevent passage through the medium, or applied without vacuum.
Follow blocking and detection procedures as indicated for the NitroBind membrane.